|
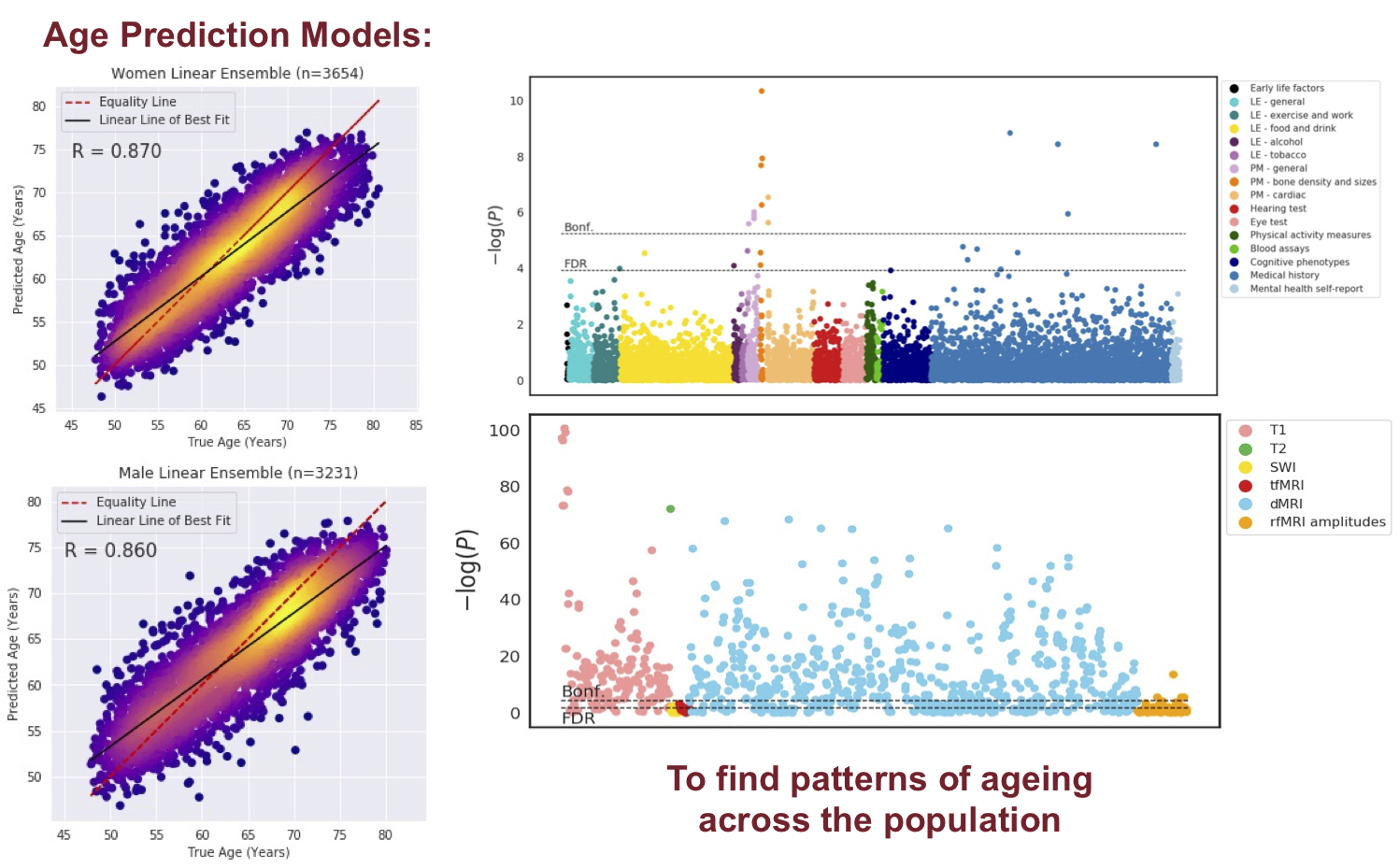
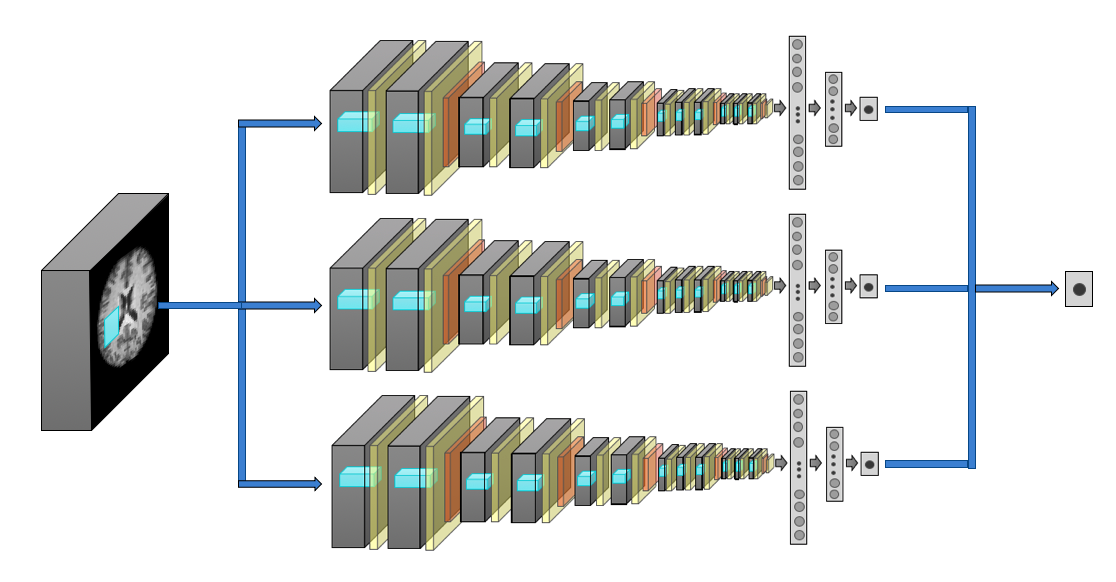
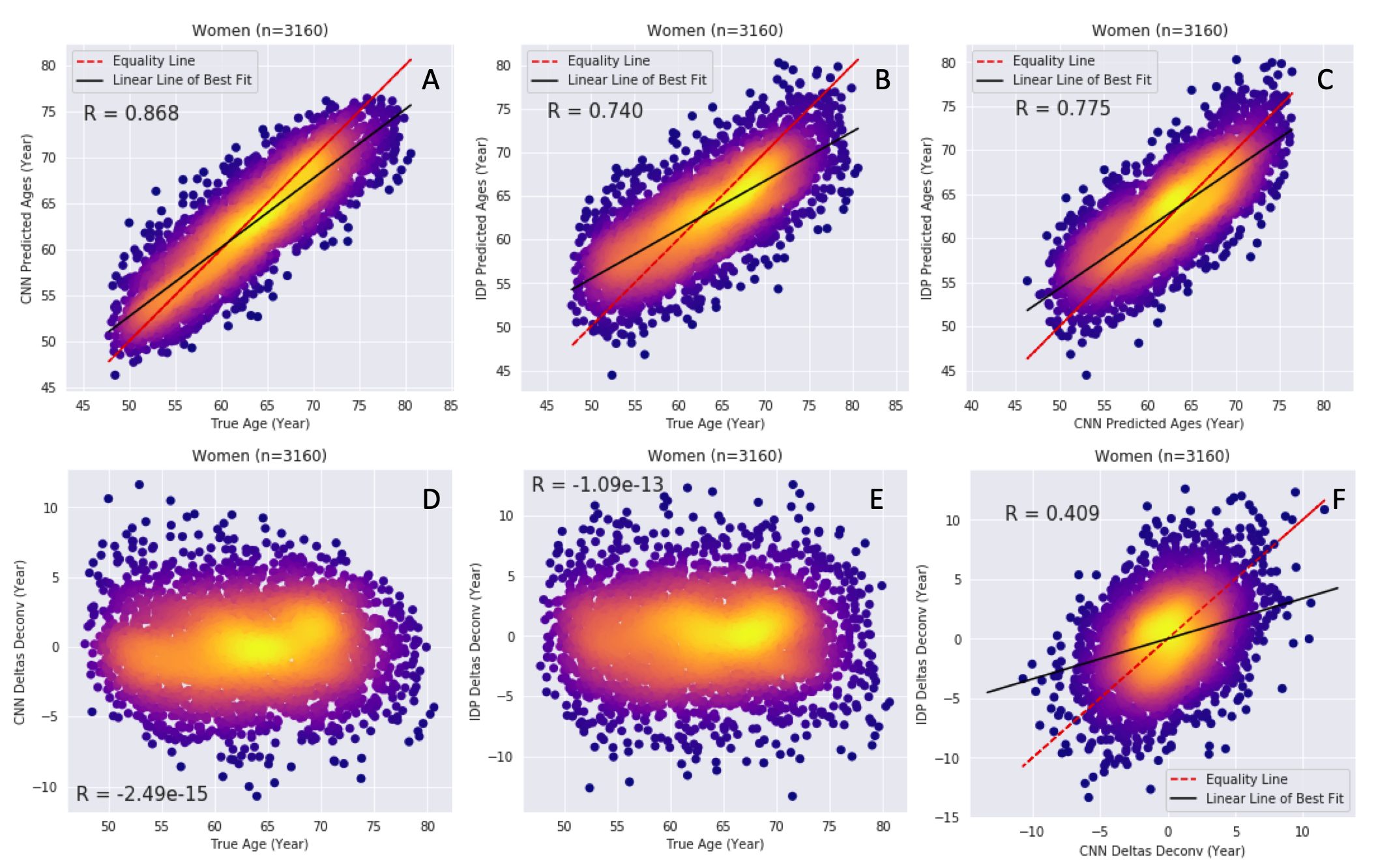
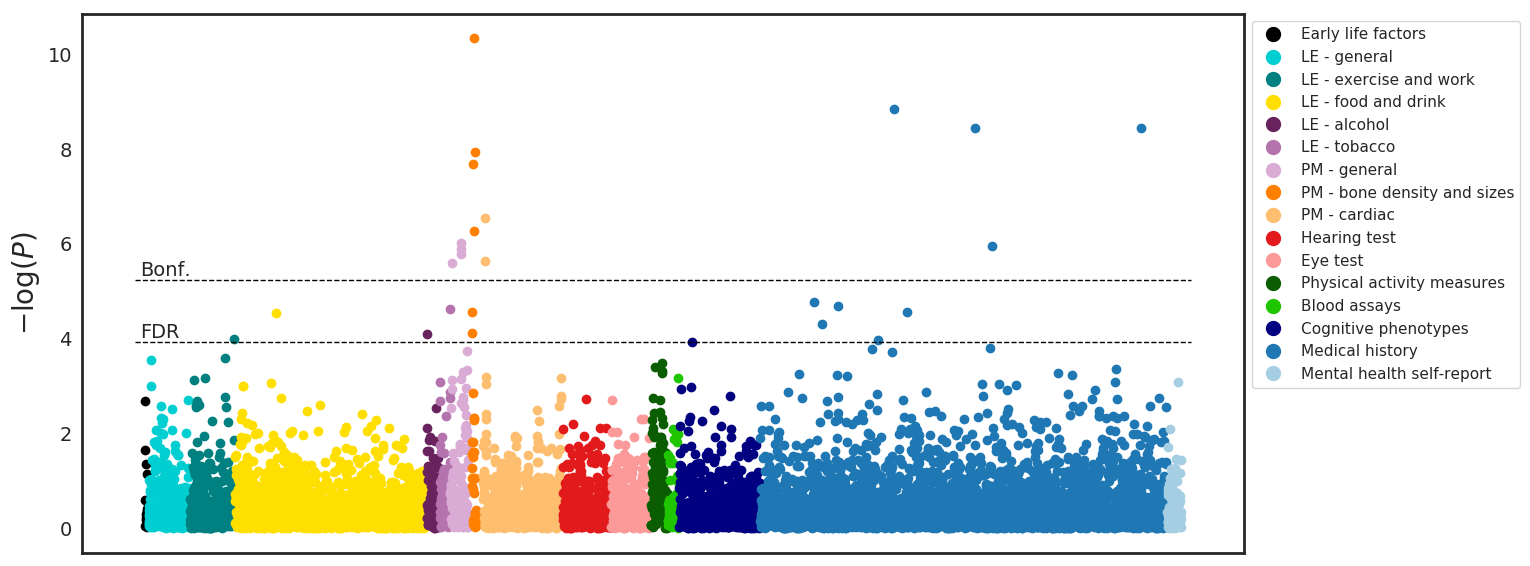
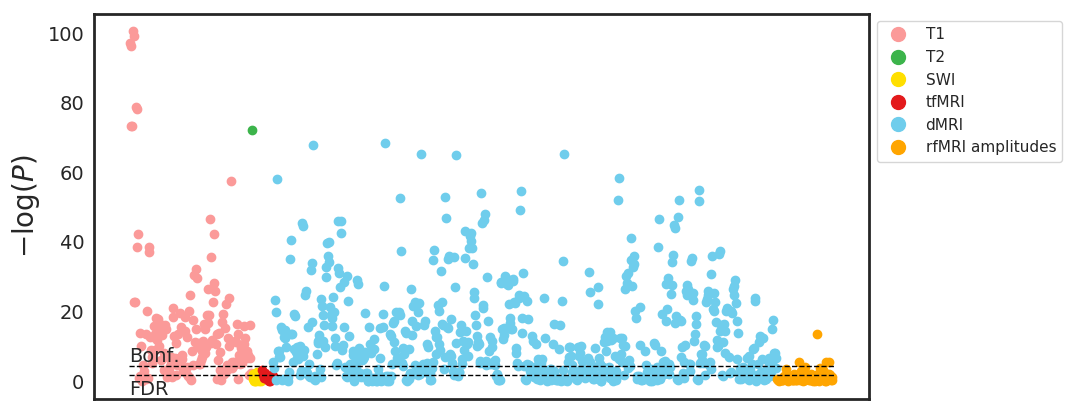
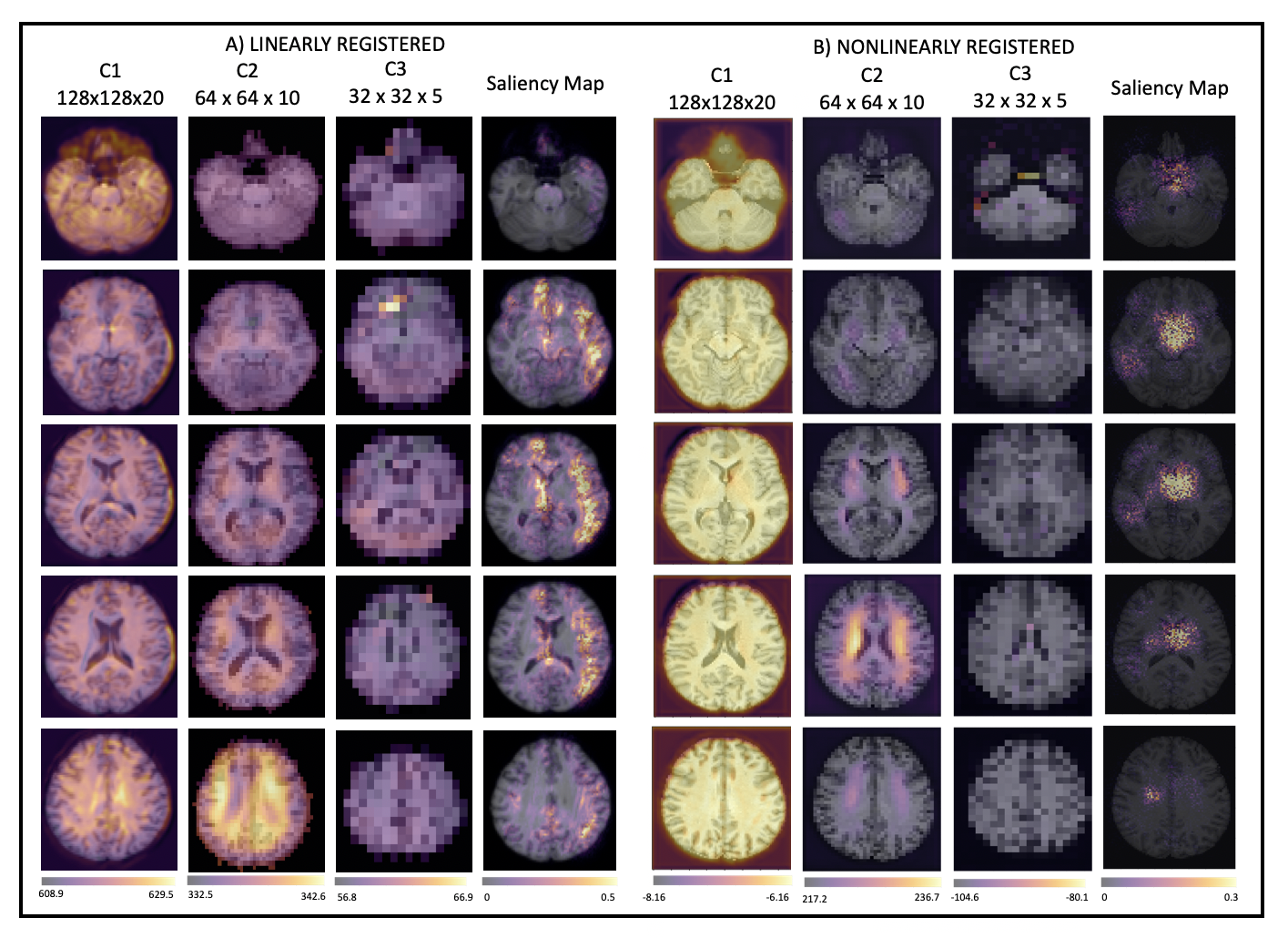
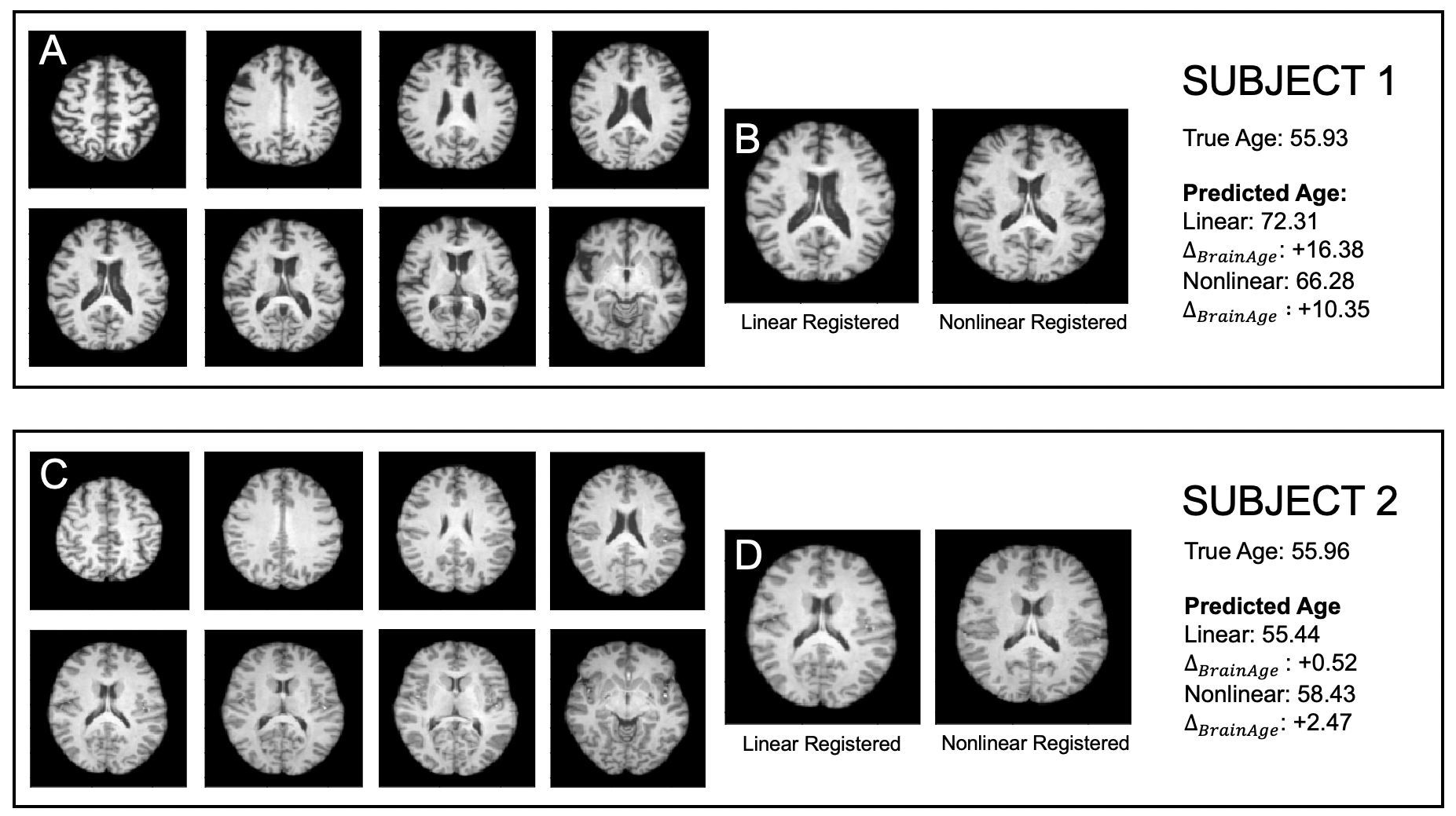
Both normal ageing and neurodegenerative diseases cause morphological changes to the brain. Age-related brain changes are subtle, nonlinear, and spatially and temporally heterogenous, both within a subject and across a population. Machine learning models are particularly suited to capture these patterns and can produce a model that is sensitive to changes of interest, despite the large variety in healthy brain appearance. In this paper, the power of convolutional neural networks (CNNs) and the rich UK Biobank dataset, the largest database currently available, are harnessed to address the problem of predicting brain age. We developed a 3D CNN architecture to predict chronological age, using a training dataset of 12,802 T1-weighted MRI images and a further 6,885 images for testing. The proposed method shows competitive performance on age prediction, but, most importantly, the CNN prediction errors correlated significantly with many clinical measurements from the UK Biobank in the female and male groups. In addition, having used images from only one imaging modality in this experiment, we examined the relationship between ΔBrainAge and the image-derived phenotypes (IDPs) from all other imaging modalities in the UK Biobank, showing correlations consistent with known patterns of ageing. Furthermore, we show that the use of nonlinearly registered images to train CNNs can lead to the network being driven by artefacts of the registration process and missing subtle indicators of ageing, limiting the clinical relevance. Due to the longitudinal aspect of the UK Biobank study, in the future it will be possible to explore whether the ΔBrainAge from models such as this network were predictive of any health outcomes.
|