Acknowledgements
ND is supported by the Engineering and Physical Sciences Research Council (EPSRC) and Medical Research Council (MRC) [grant number EP/L016052/1]. MJ is supported by the National Institute for Health Research (NIHR), Oxford Biomedical Research Centre (BRC), and this research was funded by the Wellcome Trust [215573/Z/19/Z]. The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust [203139/Z/16/Z]. AN is grateful for support from the UK Royal Academy of Engineering under the Engineering for Development Research Fellowships scheme.
This research has been conducted in part using the UK Biobank Resource under Application Number 8107. We are grateful to UK Biobank for making the data available, and to all UK Biobank study participants, who generously donated their time to make this resource possible. Analysis was carried out on the clusters at the Oxford Biomedical Research Computing (BMRC) facility and FMRIB (part of the Wellcome Centre for Integrative Neuroimaging). BMRC is a joint development between the Wellcome Centre for Human Genetics and the Big Data Institute, supported by Health Data Research UK and the NIHR Oxford Biomedical Research Centre.
The computational aspects of this research were supported by the Wellcome Trust Core Award [Grant Number 203141/Z/16/Z] and the NIHR Oxford BRC. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
The primary support for the ABIDE dataset by Adriana Di Martino was provided by the (NIMH K23MH087770) and the Leon Levy Foundation. Primary support for the work by Michael P. Milham and the INDI team was provided by gifts from Joseph P. Healy and the Stavros Niarchos Foundation to the Child Mind Institute, as well as by an NIMH award to MPM ( NIMH R03MH096321).
Data were provided in part by OASIS: Principal Investigators: T. Benzinger, D. Marcus, J. Morris; NIH P50AG00561, P30NS09857781, P01AG026276, P01AG003991, R01AG043434, UL1TR000448, R01EB009352. AV-45 doses were provided by Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly.
This template was originally made by Phillip Isola and Richard Zhang for a colorful ECCV project; the code can be found here.
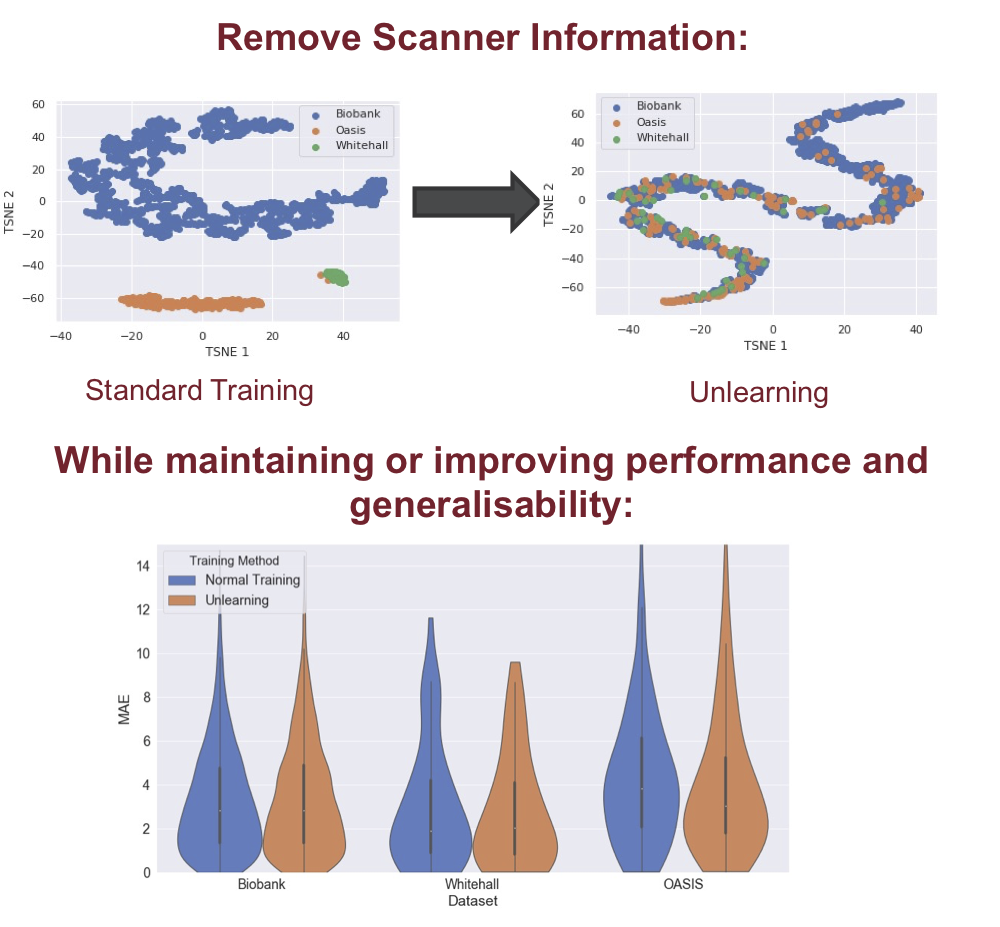
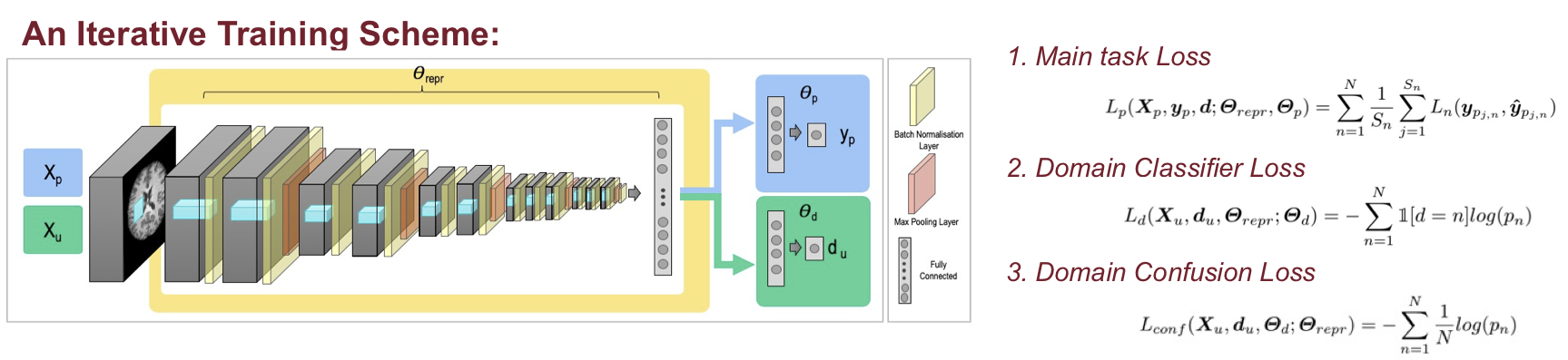
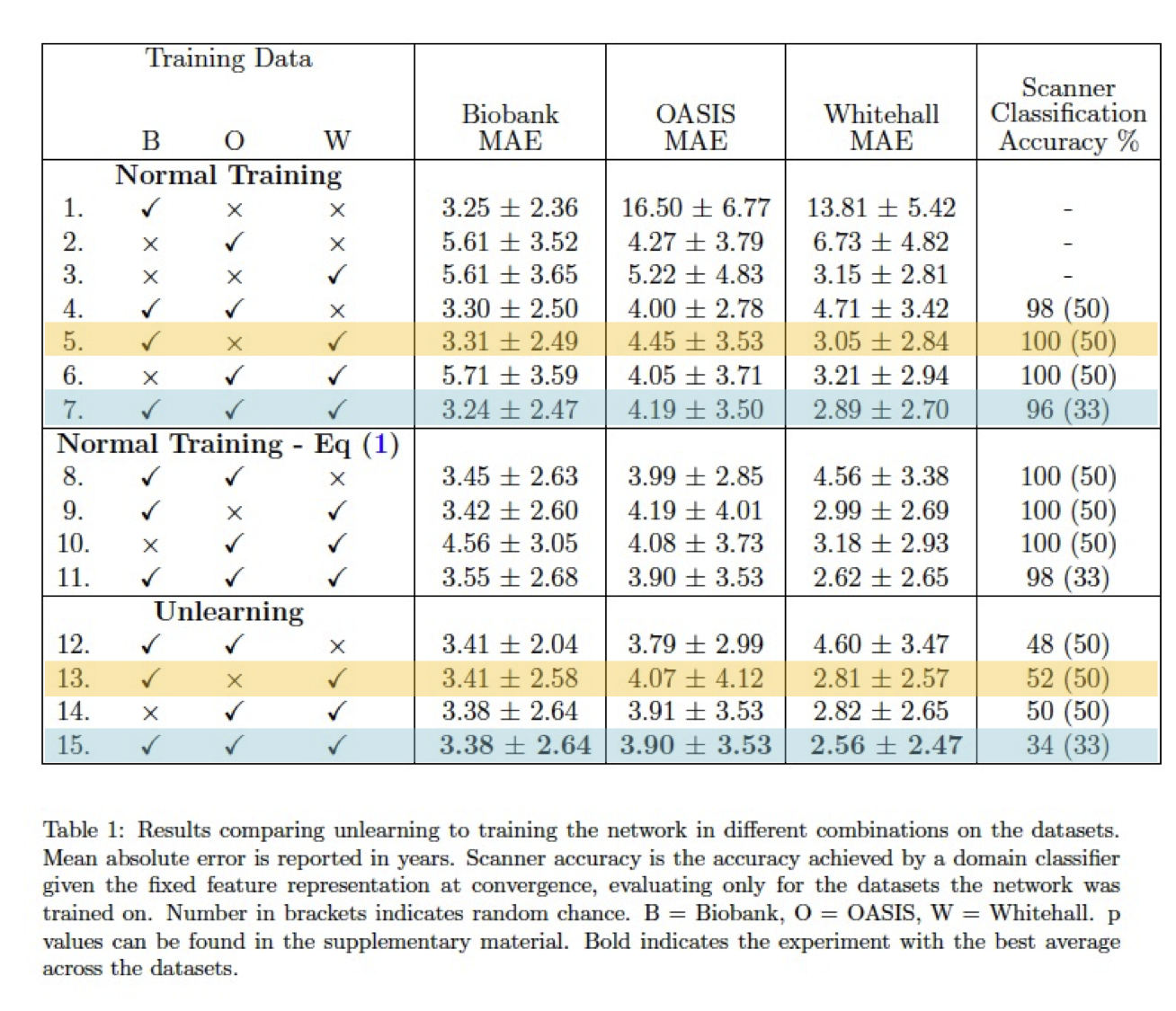
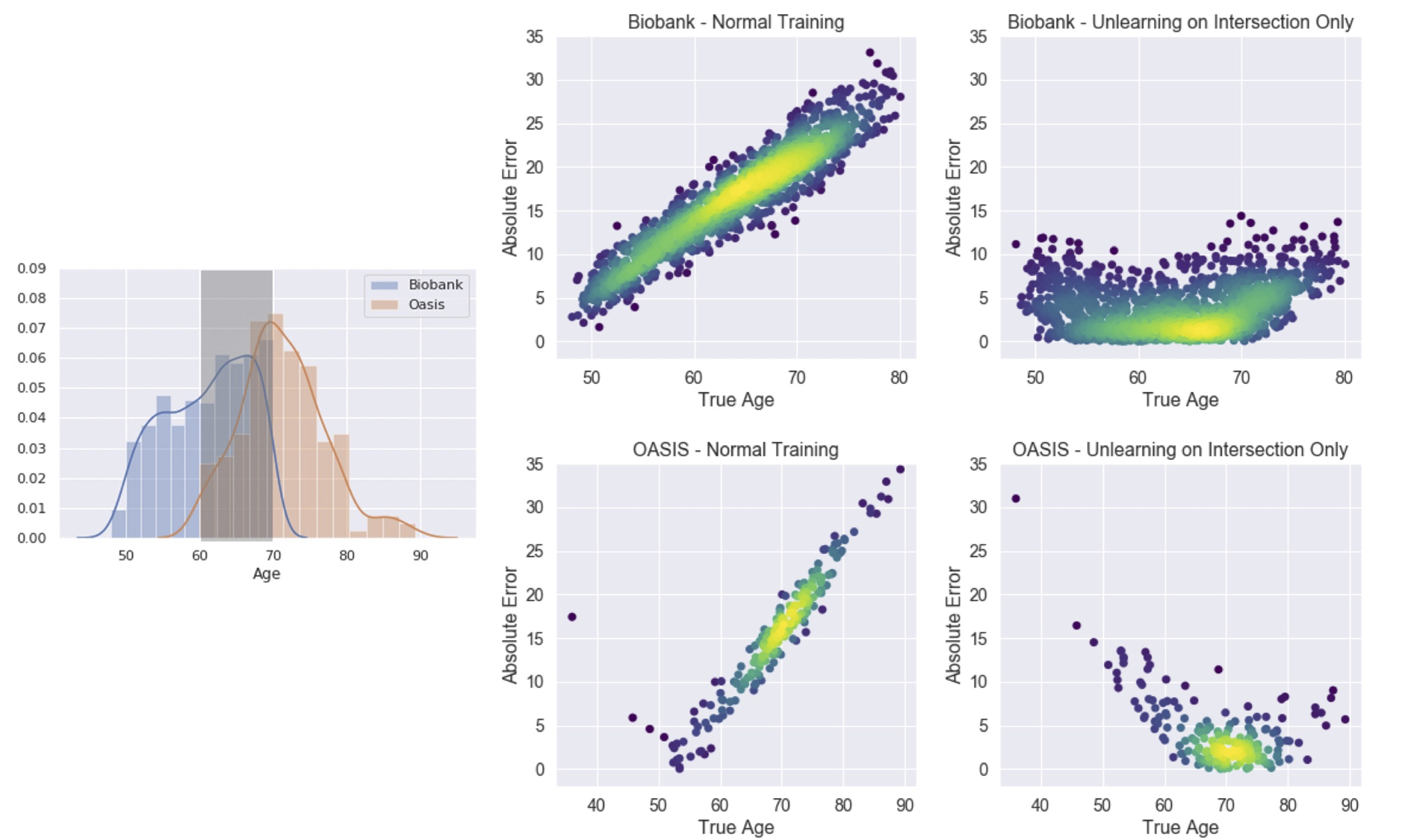
|