|
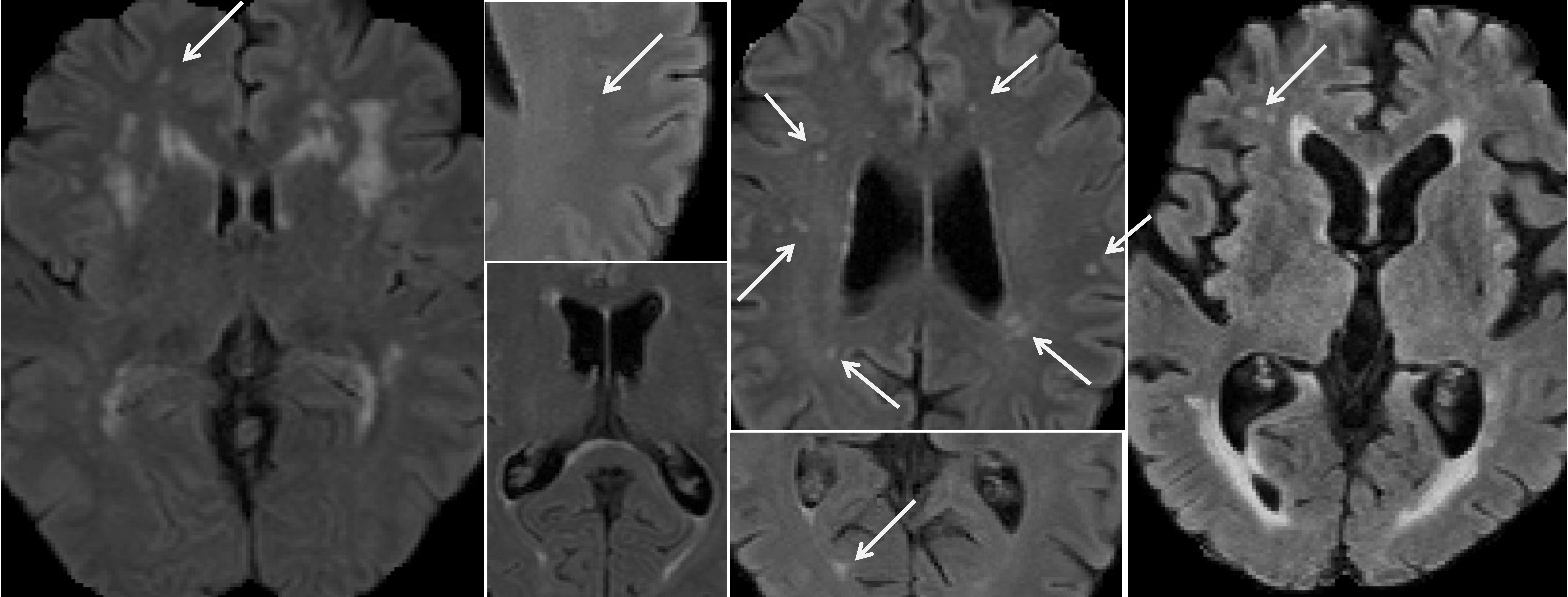
White matter hyperintensities (WMHs, or lesions) appear as
hyperintense, localized regions on T2-weighted and FLAIR
brain MR images. The heterogeneity in lesion characteristics
due to subject-level (e.g., local intensity/contrast) and
population-level (e.g., demographic, scanner-related)
variations make their segmentation highly challenging. Here,
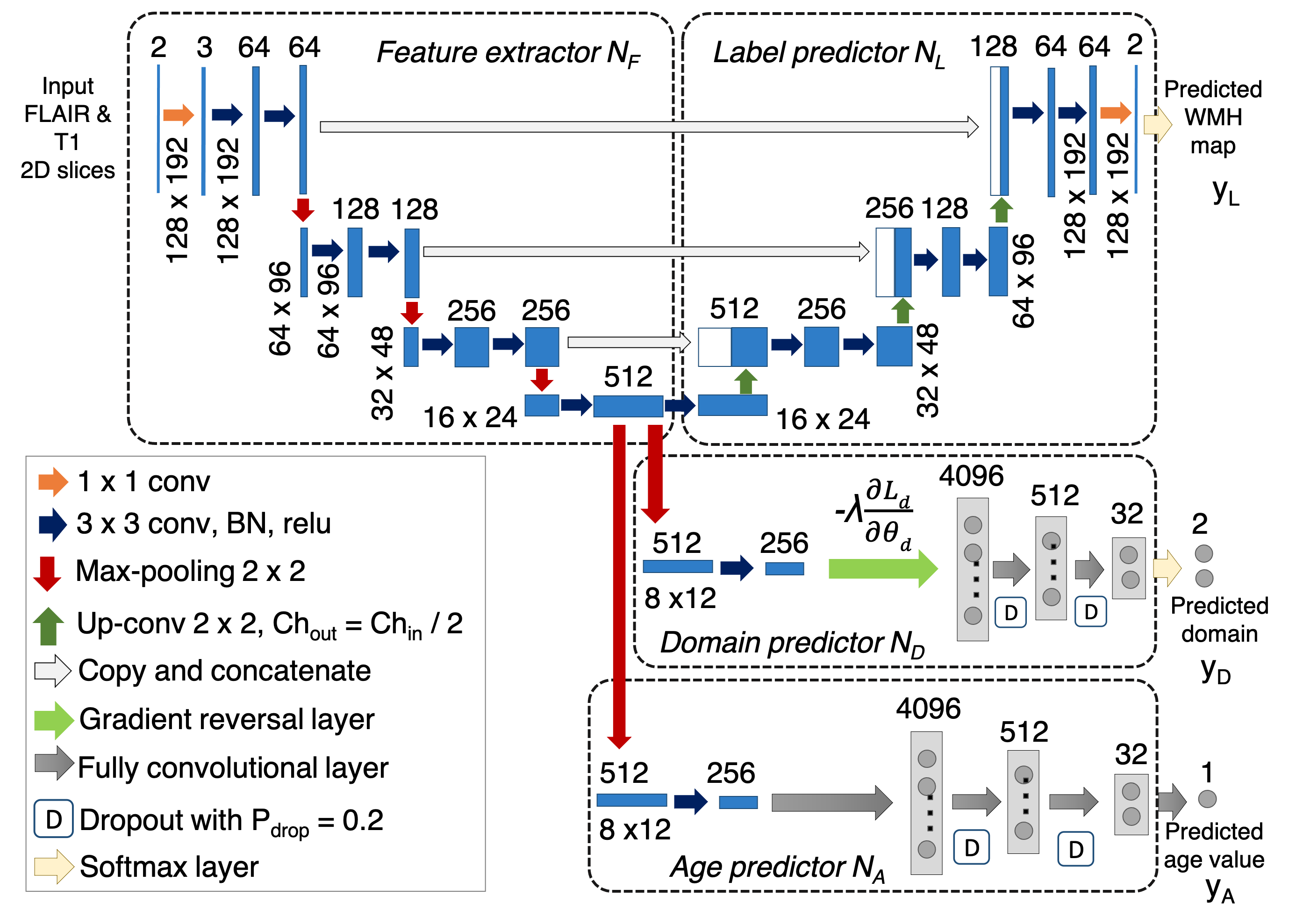
we propose a framework for adapting a state-of-the-art WMH
segmentation method with high accuracy from a small,
labeled source data (MICCAI WMH segmentation challenge
2017 training data) to a larger dataset such as the UK Biobank
without the need of additional manual training labels, using
domain adversarial training with omni-supervised learning.
Given the well-known association of WMHs with age, the
proposed method uses a multi-tasking model for learning
lesion segmentation, domain adaptation and age prediction
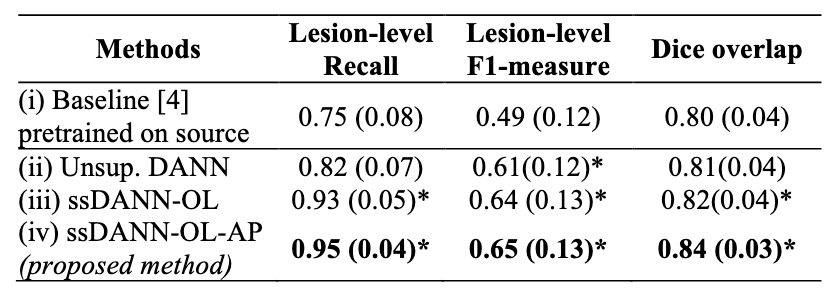
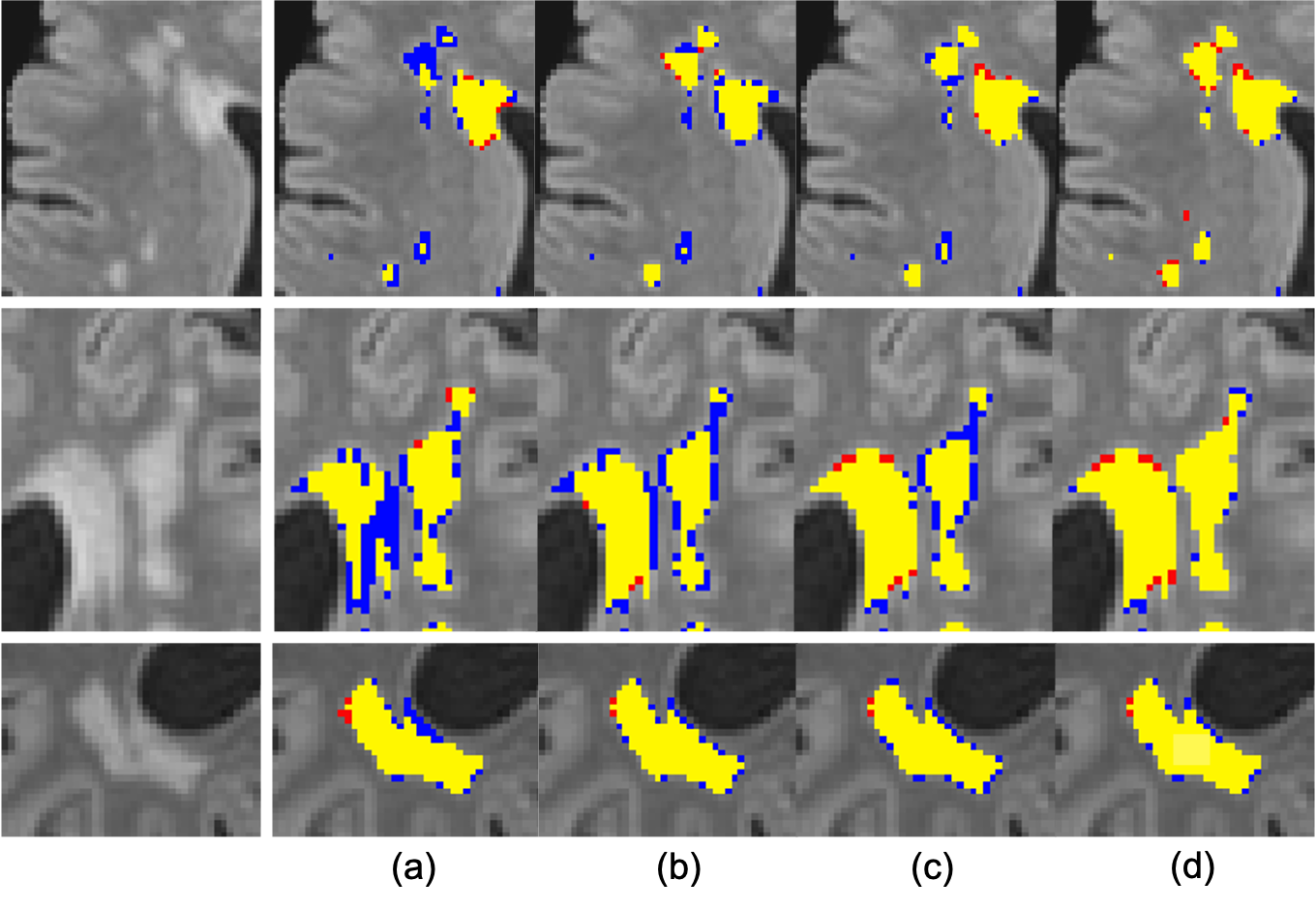
simultaneously. On a subset of the UK Biobank dataset, the
proposed method achieves a lesion-level recall, lesion-level
F1-measure and Dice overlap value of 0.95, 0.65 and 0.84
respectively, when compared to values of 0.75, 0.49 and 0.80
obtained from the pretrained state-of-the-art baseline method.
|