|
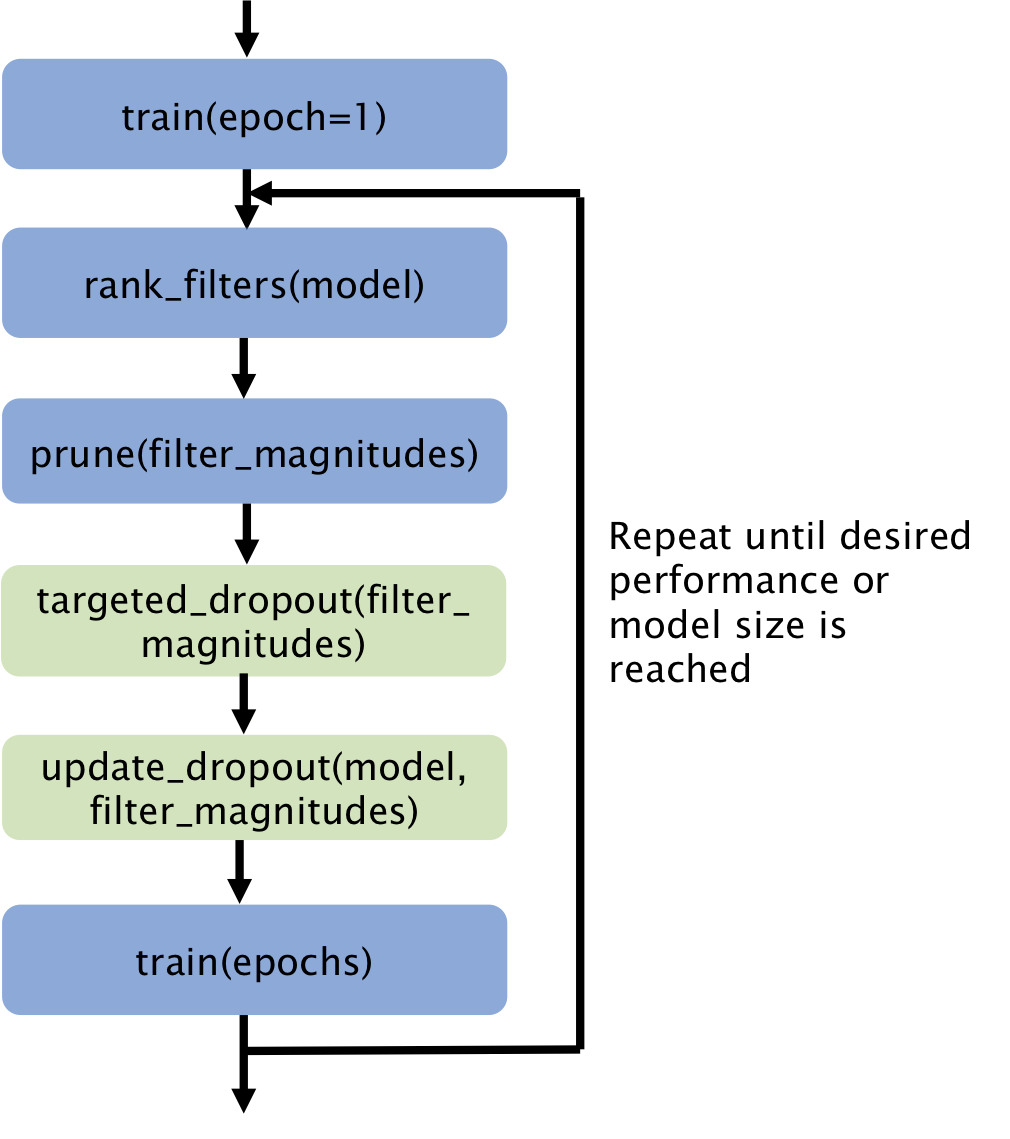
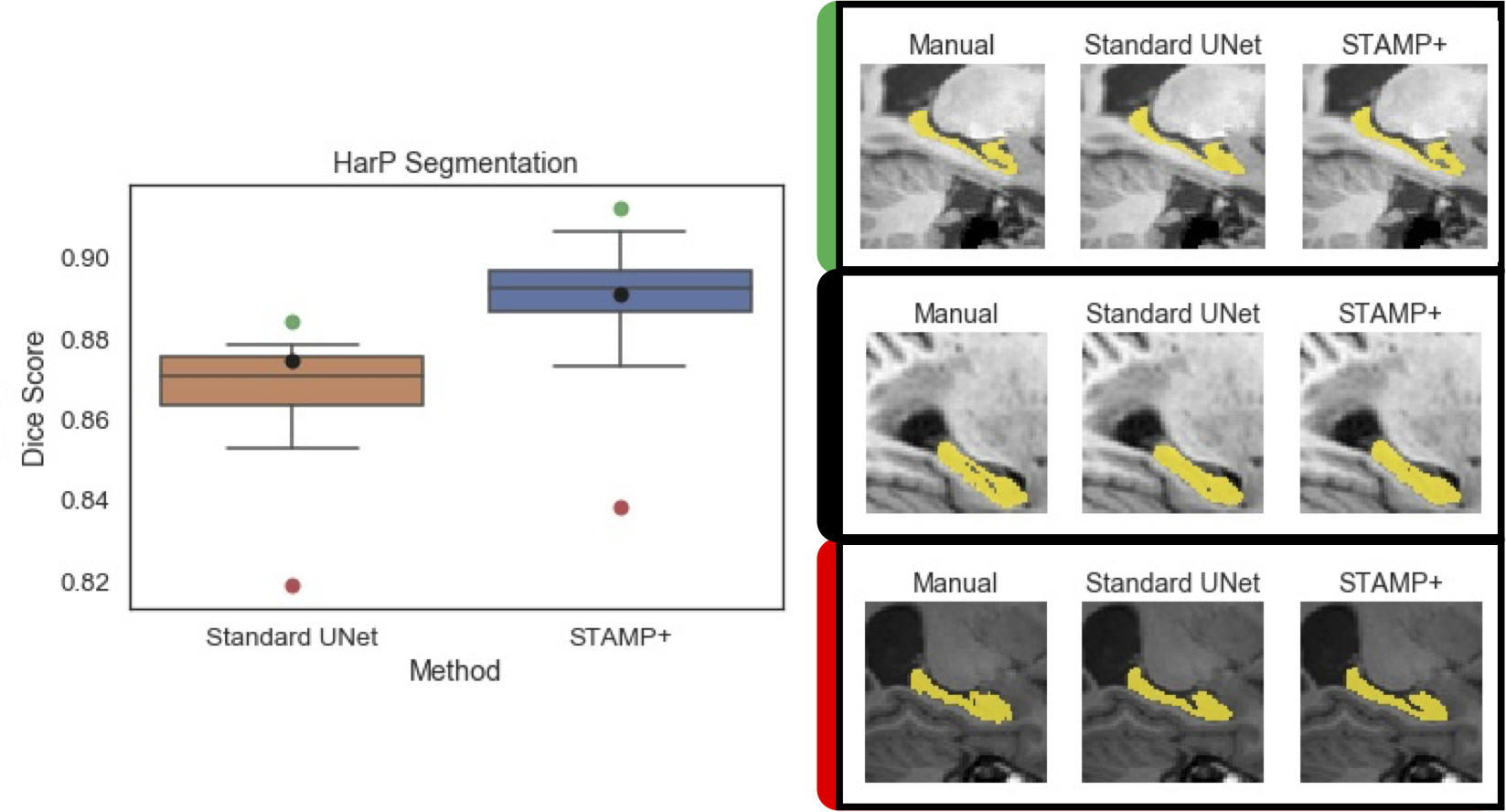
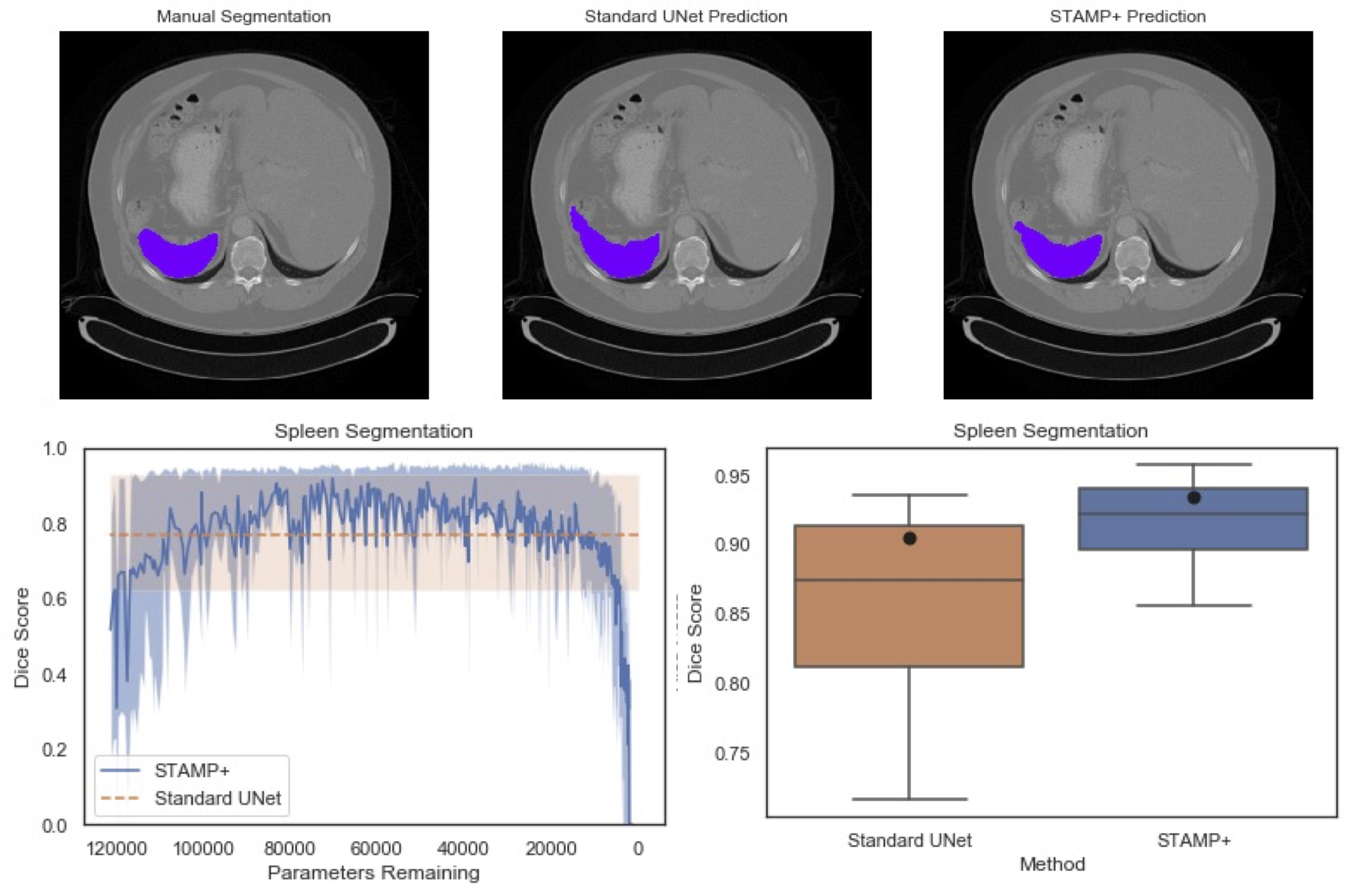
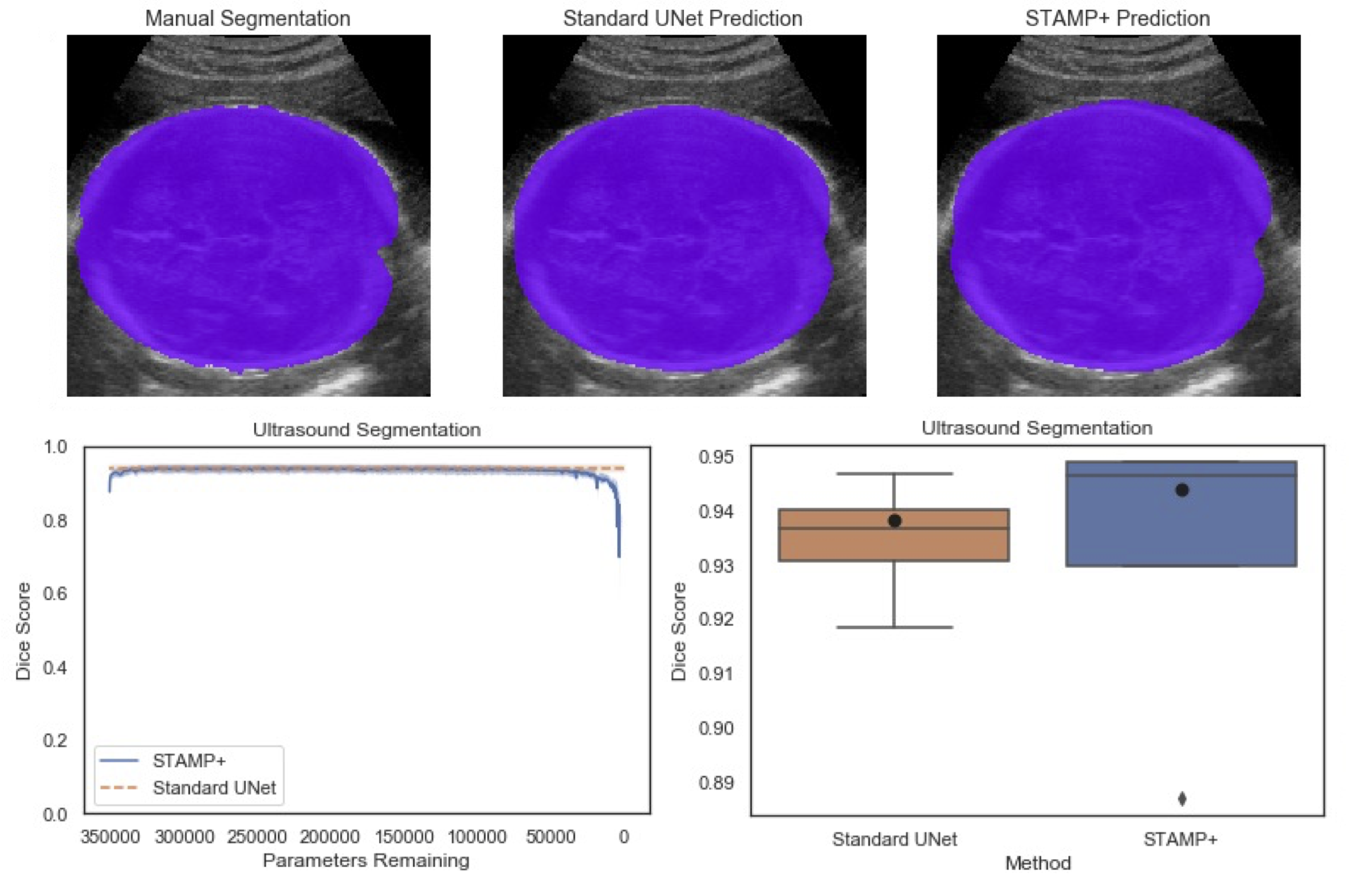
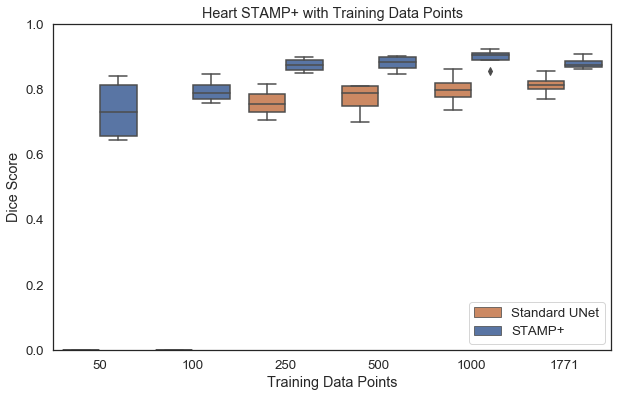
Acquisition of high quality manual annotations is vital for the development of segmentation algorithms. However, to create them we require a substantial amount of expert time and knowledge. Large numbers of labels are required to train convolutional neural networks due to the vast number of parameters that must be learned in the optimisation process. Here, we develop the STAMP algorithm to allow the simultaneous training and pruning of a UNet architecture for medical image segmentation with targeted channelwise dropout to make the network robust to the pruning. We demonstrate the technique across segmentation tasks and imaging modalities. It is then shown that, through online pruning, we are able to train networks to have much higher performance than the equivalent standard UNet models while reducing their size by more than 85% in terms of parameters. This has the potential to allow networks to be directly trained on datasets where very low numbers of labels are available. |