|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|

|
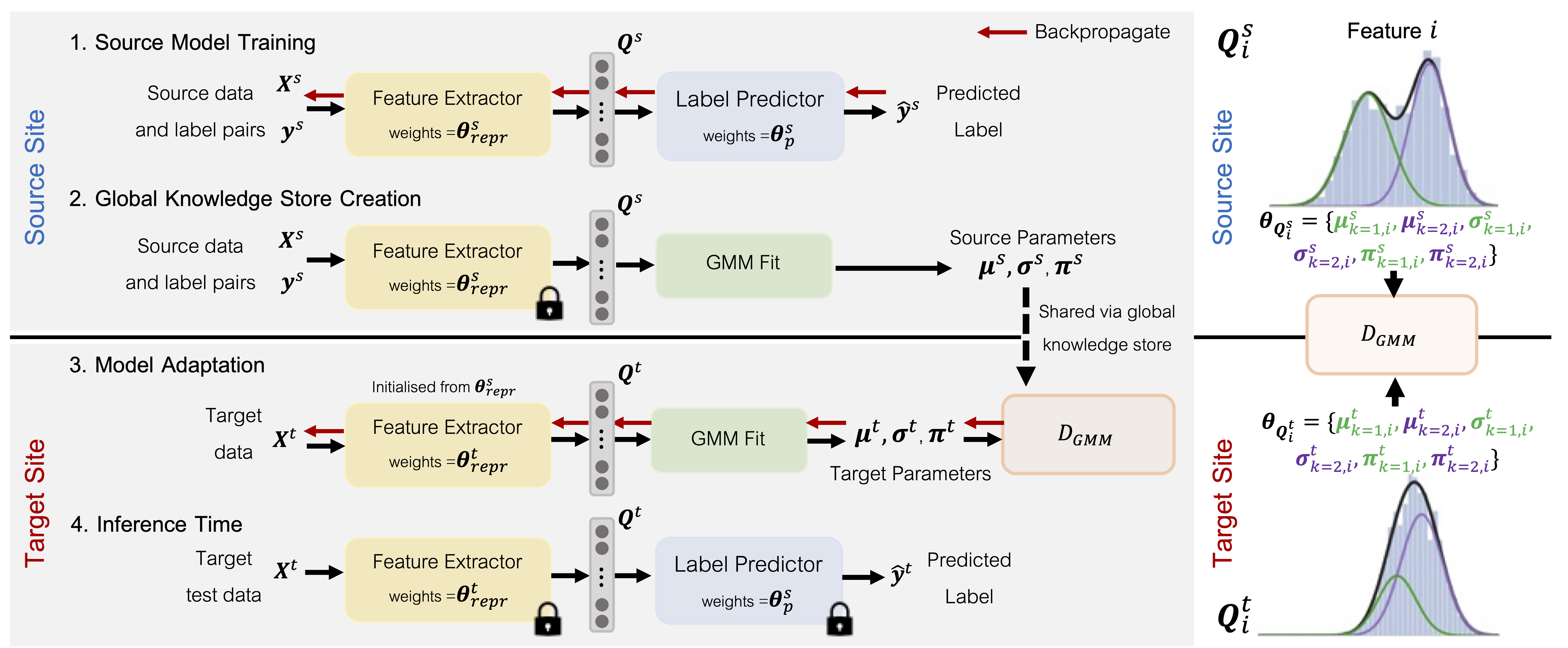
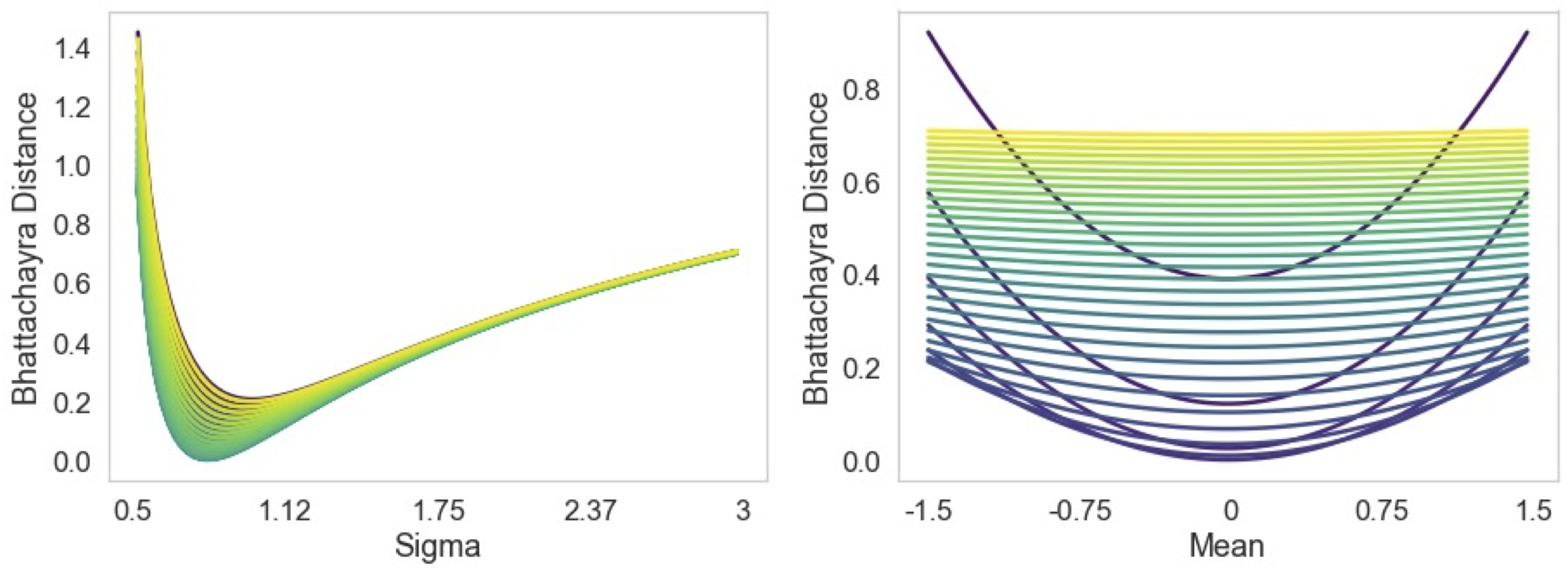
| To represent the biological variability of clinical neuroimaging populations, it is vital to be able to combine data across scanners and studies. However, different MRI scanners produce images with different characteristics, resulting in a domain shift known as the `harmonisation problem'. Additionally, neuroimaging data is inherently personal in nature, leading to data privacy concerns when sharing the data. To overcome these barriers, we propose an Unsupervised Source-Free Domain Adaptation (SFDA) method, SFHarmony. Through modelling the imaging features as a Gaussian Mixture Model and minimising an adapted Bhattacharyya distance between the source and target features, we can create a model that performs well for the target data whilst having a shared feature representation across the data domains, without needing access to the source data for adaptation or target labels. We demonstrate the performance of our method on simulated and real domain shifts, showing that the approach is applicable to classification, segmentation and regression tasks, requiring no changes to the algorithm. Our method outperforms existing SFDA approaches across a range of realistic data scenarios, demonstrating the potential utility of our approach for MRI harmonisation and general SFDA problems. |
|
| We explore an unsupervised DA setting where only the source model, instead of the source data, is pro- vided to the unlabelled target domain for harmonisation, known as Source Free Domain Adaptation (SFDA). This setting inherently protects individual privacy, whilst allow- ing the efficient incorporation of new sites without requiring target labels. We propose a simple yet effective solution, termed SFHarmony, which aims to match feature embed- dings from the source and target, through characterising the embeddings as a Gaussian Mixture model (GMM) and the use of a modified Bhattacharyya distance. We demonstrate the approach for classification, segmentation and regression tasks. |
| Our contributions are as follows: |
| - We propose a new method for SFDA, SFHarmony, based on aligning feature embeddings, utilising a modified Bhattacharyya distance, requiring no changes to source training; |
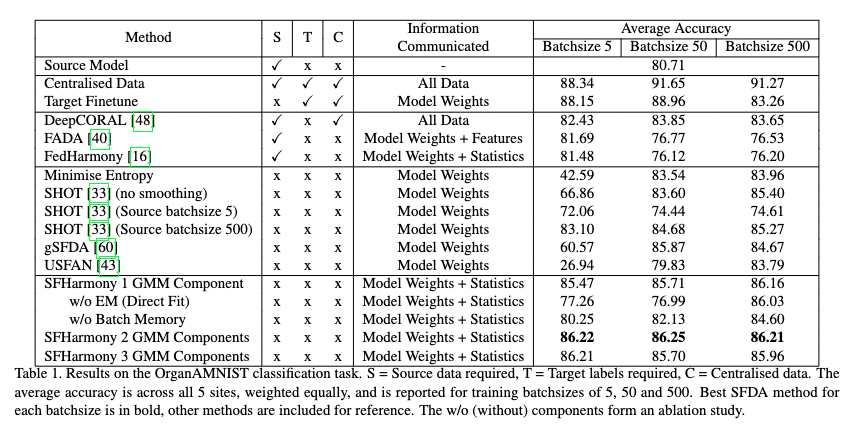
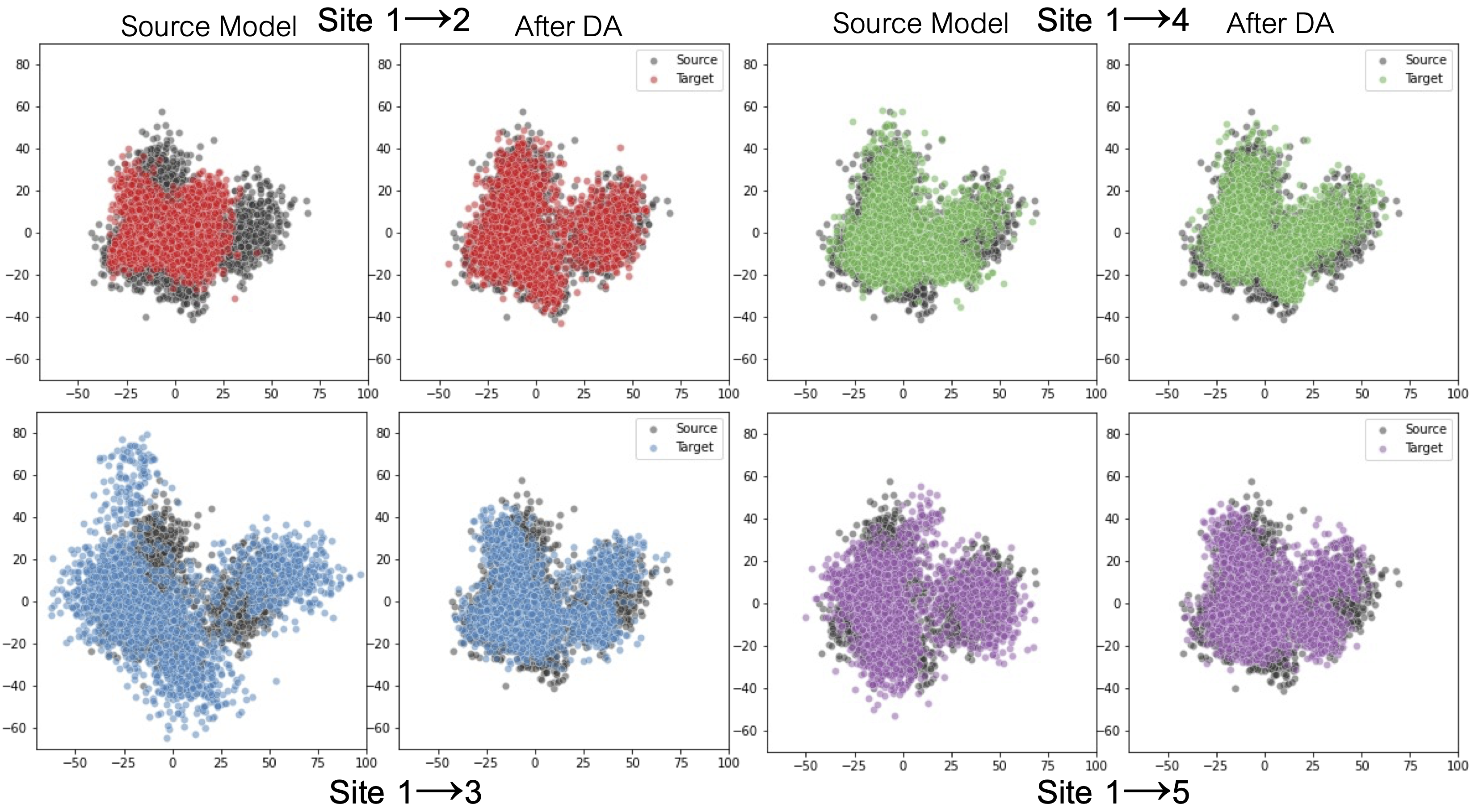
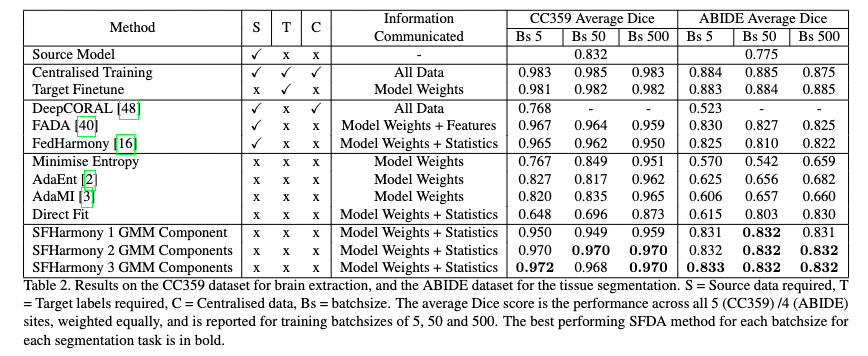
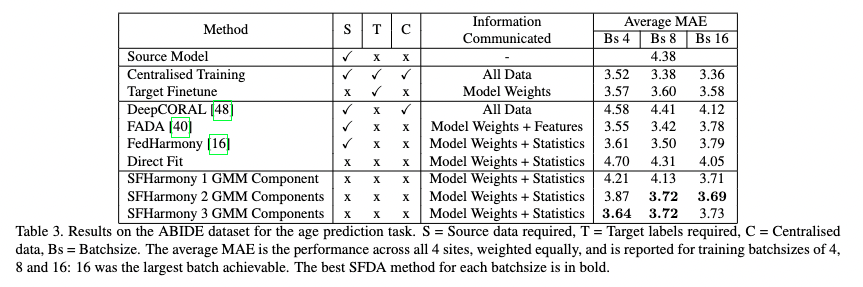
| - We demonstrate the method’s applicability to classification, segmentation and regression tasks, and show that the approach outperforms existing SFDA methods for domain shifts experienced when working with neuroimaging data; |
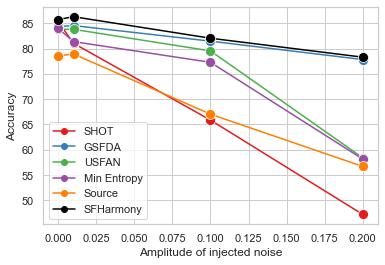
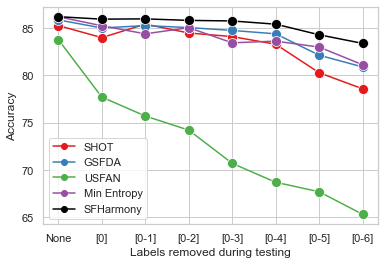
| - We demonstrate the robustness of the method to additional challenges likely to be faced when working with real world imaging data: differential privacy and label imbalance. |
Conclusion |
Acknowledgements |