|
The delineation of regions in medical images - segmentation - is a key task across modalities and organs. Methods developed need to produce segmentations that are biologically meaningful across scanners and imaging protocols. Home / Harmonisation / Segmentation / Translation / Privacy / Explainable AI |
|
|
|
|
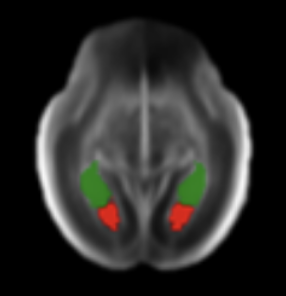
Joshua Omolegan, Pak Hei Yeung, Madeleine K Wyburd, Linde Hesse, Monique Haak, Intergrowth Consortium, Ana IL Namburete, Nicola K Dinsdale ISBI 2025 Paper / Code Test time adaptation for robust subcortical segmentation in fetal ultrasound |

|
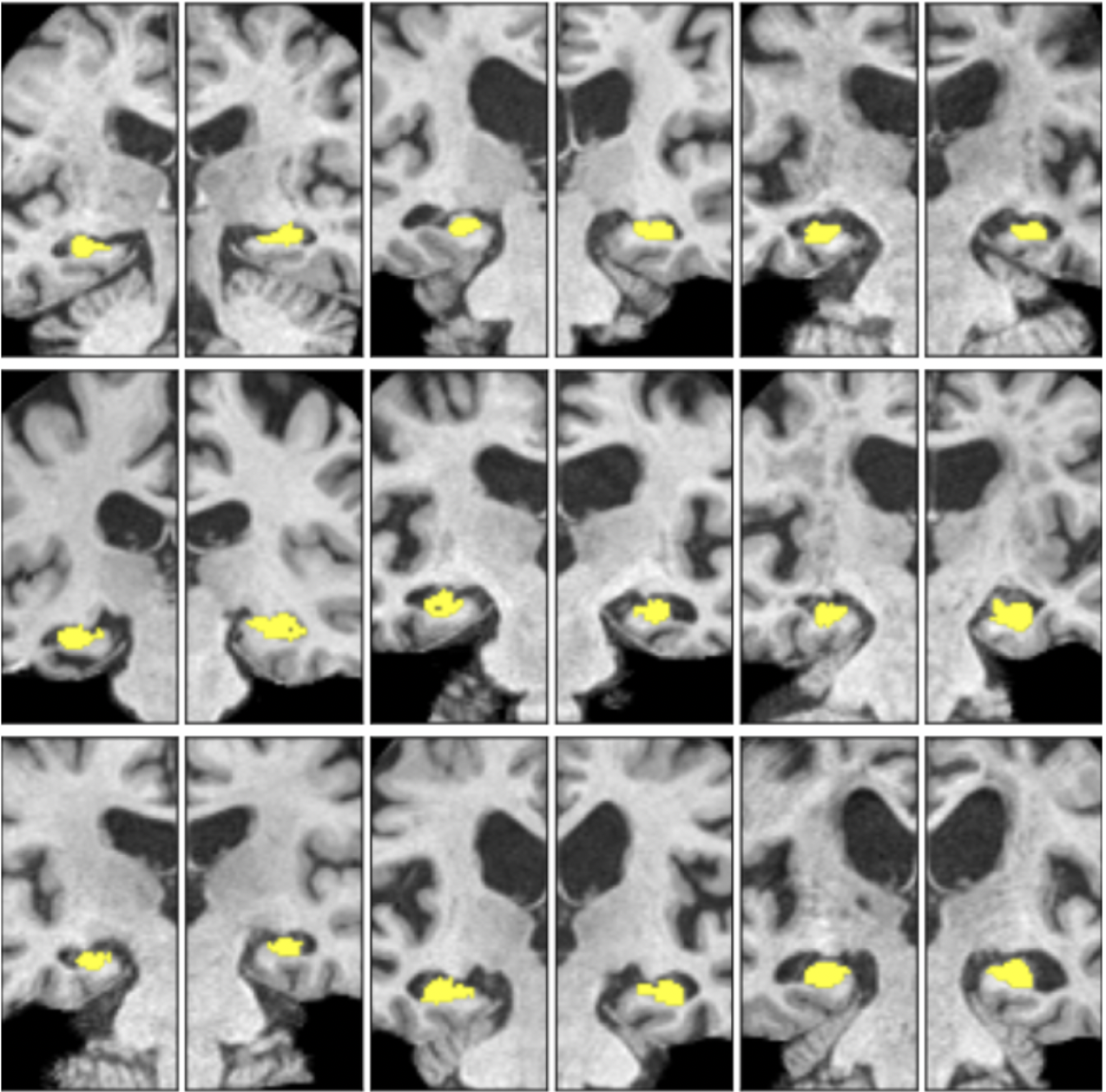
Vaanathi Sundaresan, Nicola K Dinsdale Low Field Pediatric Brain Magnetic Resonance Image Segmentation and Quality Assurance Challenge, 2024 Paper / Code Winner of LISA Challenge 2024 @ MICCAI 2024 |
|
Hoda Kalabizadeh, Ludovica Griffanti, Pak-Hei Yeung, Natalie Voets, Grace Gillis, Clare E Mackay, Ana IL Namburete, Nicola K Dinsdale*, Konstantinos Kamnitsas* Machine Learning in Clinical Neuroimaging, 2024 Paper We investigated the performance of segmentation models trained on research data that were style-transferred to resemble clinical scans. Our results highlighted the importance of intensity normalisation methods in MRI segmentation, and their relation to domain shift and style-transfer. |

|
Madeleine K Wyburd, Nicola K Dinsdale , Ana IL Namburete, Mark Jenkinson Medical Image Analysis 2024 Paper / Code Our model, TEDS-Net, generates anatomically plausible segmentations through deforming a prior shape with the same topology as the anatomy of interest. |
|
Nicola K Dinsdale, Mark Jenkinson, Ana IL Namburete bioRxiv, 2024 Project Page / Paper / Code We, therefore, propose UniFed, a unified federated harmoni- sation framework, which enables three key processes to be completed: 1) the training of a federated partially labelled harmonisation network, 2) the selection of the most appropriate pretrained model for a new unseen site, and 3) the incorporation of a new site into the harmonised federation. |
|
Nicola K Dinsdale , Mark Jenkinson, Ana IL Namburete Medical Image Analysis, 2022 Project Page / Paper / Code Development of an algorithm to enable simultaneous training and pruning, enabling segmentation in low data domains. Also introduces Targeted Dropout to stabilise the pruning. |
|
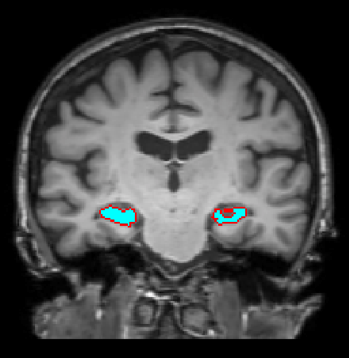
Vaanathi Sundaresan, Nicola K Dinsdale, Ludovica Griffanti, Mark Jenkinson, ISBI 2022 [Oral Presentation] Project Page / Paper / Code Exploring the use of omni-supervised learning for white matter hyperintensity segmentation, using age prediction as the selection criteria. Leads to significant increase in sensitivity especially for small lesions. |
|
Erica Balboni, Luca Nocetti, Chiara Carbone, Nicola K Dinsdale , Maurilio Genovese, Gabriele Guidi, Marcella Malagoli, Annalisa Chiari, Ana IL Namburete, Mark Jenkinson, Giovanna Zamboni, Human Brain Mapping 2022 Paper / Code Exploration of the use of my SWANS algorithm for clinical data, across disease groups - SWANS first presented at MICCAI 2019 |

|
Madeleine K Wyburd, Nicola K Dinsdale , Ana IL Namburete, Mark Jenkinson MICCAI, 2021 [Early Acceptance] Project Page / Paper / Code Novel segmentation method guaranteeing accurate topology. |

|
Vaanathi Sundaresan, Giovanna Zamboni, Nicola K Dinsdale , Peter Rothwell, Ludovica Griffanti, Mark Jenkinson Medical Image Analysis 2021 Paper / Code Comparison of DA methods for white matter hyperintensity segmentation, comparing methods including my proposed method: paper. |
|
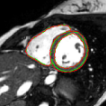
Jorge Corral Acero, Vaanathi Sundaresan, Nicola K Dinsdale , Vicente Grau, Mark Jenkinson STACOM 2020 - 6th place Paper Segmentation of cardiac MRI across sites, using the method proposed in my paper |

|
Nicola K Dinsdale , Mark Jenkinson, Ana IL Namburete MICCAI, 2019 [Early Acceptance] Paper / Code Novel segmentation method based on a spatial transformer network. |
The template of this webpage is from source code.